Part 1: How to design sgRNA
Finding an sgRNA that effectively causes loss of gene function is seemingly the most important step in the entire complex process of gene KO. Because if the sg design is not good (or you can just call it unlucky for fun), then after a series of days of hard work, a lot of effort and money spent on chemicals, but then running the website will be protein. of that gene is still inert, that gene still functions normally, so how can we study the role of that gene in the cell for the purpose of deleting the gene, right?
Therefore, usually most researchers will choose at least 5 sgRNAs for 1 target gene. For me, I usually choose 10 sgRNAs for 1 gene. Here is an example for everyone to observe, the article has Posted a few months ago, if anyone is interested, please message me and I will send you the full article, but this article requires you to buy it to read it.
Figure 1: sgCTRL are sgRNAs that also contain 20 RNAs but do not target any genes in the human genome.
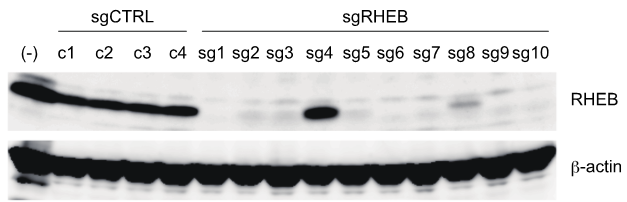
gRHEB.1 to sgRHEB.10 are 10sg that I selected from the available library system. These sg have been selected using bioiformatic and available smart computer software.
Figure 1: shows us that all 4 control sg completely do not lose the band of the RHEB protein, proving that the gene that regulates the trait for the RHEB protein is completely normal and does not affect the structure and function of the RHEB gene. chief. (RHEB band about 20 Kda)
Meanwhile, 10sgRHEB has up to 9 sg loss of band of RHEB, only sg4, the band of RHEB, is still intact, proving that sg4 is located on an unimportant exon, after gene KO, it does not lose function at all. function of the RHEB gene.
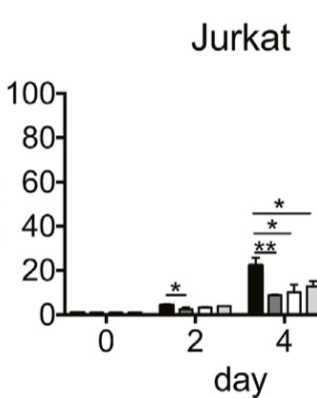
I chose 3 sg out of 9 sg with the effectiveness shown in photo 1 to continue doing further experiments on many different cancer cell lines to study the role of the RHEB gene: sg5, sg6, sg9 : photo 2
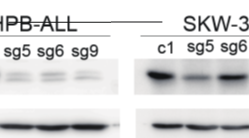
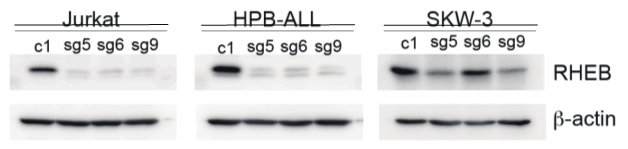
Figure 3: understanding the mechanism of RHEB in the RHEB gene KO model
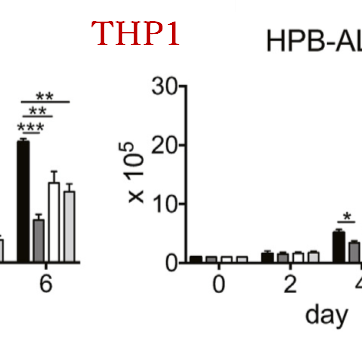
After KOing the RHEB gene, the KO cell line clearly changes the mechanisms related to S6K, 4E-BP (mechanism of carcinogenesis) as shown in the wb experiment. Actin is the control protein for the RHEB gene KO experiment.
Tools for designing sg for target genes
General overview website.
https://en.wikipedia.org/wiki/CRISPR/Cas_tools
https://zlab.bio/guide-design-resources
The site I often use to design sgRNA
https://www.synthego.com/products/bioinformatics/crispr-design-tool
http://chopchop.cbu.uib.no
https://www.thermofisher.com/us/en/home/life-science/genome-editing/gene...
Another day, I will show you how to design sgRNA by hand. It seems that it will be easier and deeper to understand than the case where everything is thrown on the computer and the software runs automatically like this.
The simplest way is to have a sgRNA library for 18,166 genes in the human genome, find it and take it out. It's the simplest and most effective and saves a lot of neurons.